Genome-Wide Association Studies: Manhattan Plot Excellence in Genomics Research
Master Manhattan plot creation for GWAS and genomics research through real examples from Nature Genetics and leading journals. Learn significance thresholds, genomic coordinate mapping, and interpretation.
Throughout my career specializing in genome-wide association studies and population genetics, I have consistently observed Manhattan plots serving as the definitive visualization for revealing genome-wide patterns of genetic association that guide disease mechanism discovery and therapeutic target identification. Their unique ability to simultaneously display statistical significance across all chromosomes while preserving genomic coordinate information makes them indispensable for studies where identifying disease-associated loci drives precision medicine development and evolutionary insight generation.
Application Scenarios Across Genomics Research
In my extensive analysis of Manhattan plot implementations across major genetics journals, I observe sophisticated application patterns that demonstrate both analytical rigor and biological discovery potential:
• Complex Disease Genetics and GWAS: Publications in Nature Genetics and Cell consistently feature Manhattan plots for presenting genome-wide association results across diverse diseases, phenotypes, and population cohorts. I have reviewed countless GWAS studies where Manhattan plots serve as the primary tool for identifying disease susceptibility loci while simultaneously revealing polygenic architecture patterns and population-specific genetic effects. The disease genetics context particularly benefits from Manhattan plot visualization, where researchers must balance genome-wide significance thresholds with biological plausibility to identify genuine disease associations while controlling false discovery rates across millions of genetic variants.
• Pharmacogenomics and Drug Response Studies: Clinical pharmacology research publications routinely employ Manhattan plots for presenting drug response associations, adverse event susceptibility loci, and treatment efficacy variant identification across diverse patient populations and therapeutic interventions. I observe these visualizations proving essential for identifying pharmacogenetic markers, revealing population-specific drug response patterns, and demonstrating genetic factors that influence therapeutic windows and safety profiles. The pharmacogenomics context requires sophisticated population stratification and ancestry adjustment that influences Manhattan plot interpretation and clinical translation potential.
• Evolutionary Genetics and Population Studies: Population genetics research frequently utilizes Manhattan plots for presenting selection signatures, population differentiation patterns, and demographic history inference across diverse human populations and model organisms. In my review experience, these visualizations excel at revealing genomic regions under selection, identifying population-specific adaptations, and demonstrating evolutionary constraints that shape genetic diversity patterns. The evolutionary context often requires sophisticated statistical models that account for population structure, demographic history, and linkage disequilibrium patterns that influence Manhattan plot interpretation.
• Molecular QTL Mapping and Functional Genomics: Functional genomics publications consistently employ Manhattan plots for presenting expression quantitative trait loci (eQTL), protein QTL, and methylation QTL results across different tissues, developmental stages, and environmental conditions. I have analyzed numerous QTL studies where Manhattan plots reveal regulatory variant effects, identify tissue-specific genetic regulation patterns, and demonstrate environmental interaction effects that influence molecular phenotype variation and disease susceptibility mechanisms.
Strengths and Limitations of Manhattan Plot Visualization
Through my extensive experience implementing Manhattan plots across diverse genomics research contexts, I have identified both the remarkable analytical capabilities and inherent challenges of this visualization approach:
Key Strengths
• Genome-Wide Pattern Recognition and Locus Identification: Manhattan plots excel at revealing genome-wide association patterns while enabling immediate identification of significant loci and assessment of overall polygenic architecture across entire genomes or chromosomal regions. During my GWAS analyses, I consistently rely on Manhattan plots to identify novel disease loci, assess replication patterns across studies, and evaluate genome-wide significance distributions that inform statistical power and study design decisions. The chromosomal organization provides intuitive genetic mapping while maintaining statistical rigor necessary for genetic discovery and clinical translation.
• Multiple Testing Control and Statistical Rigor: Superior integration with genome-wide significance thresholds enables Manhattan plots to appropriately control false discovery rates while highlighting genuinely significant associations that warrant follow-up functional validation and clinical investigation. I have observed how well-designed Manhattan plots consistently incorporate appropriate Bonferroni correction thresholds, permutation-based significance assessment, and multiple testing considerations that maintain discovery reliability while enabling identification of novel genetic associations with clinical relevance.
• Population Structure and Ancestry Visualization: Advanced Manhattan plot implementations can reveal population stratification effects, ancestry-specific associations, and demographic confounding through systematic patterns of significance distribution and genomic inflation assessment. In my population genetics research, I frequently employ Manhattan plots that incorporate population structure corrections, ancestry-specific analyses, and demographic modeling results that enable identification of genuine genetic associations while controlling for population history and migration pattern confounders.
Primary Limitations
• Linkage Disequilibrium and Causal Variant Identification: Manhattan plots cannot distinguish between causal variants and linked markers in regions of strong linkage disequilibrium, potentially leading to imprecise localization of functional variants and overestimation of independent association signals within genomic regions. I frequently encounter situations during manuscript reviews where Manhattan plot peaks represent single causal variants with multiple correlated signals rather than independent genetic effects, requiring sophisticated fine-mapping approaches and functional validation studies to identify genuine causal mechanisms.
• Effect Size Communication and Clinical Relevance: While Manhattan plots effectively communicate statistical significance, they provide limited information about effect sizes, clinical relevance, and population attributable risk that are critical for assessing genetic variant importance and therapeutic target potential. During collaborative clinical research, I often observe how statistically significant Manhattan plot peaks may represent variants with minimal clinical impact, emphasizing the importance of integrating effect size assessment with statistical significance evaluation for clinical translation and therapeutic development decisions.
• Population Generalizability and Ancestry Bias: Manhattan plot interpretation may be limited by population-specific linkage disequilibrium patterns, ancestry-specific effect sizes, and demographic history influences that affect generalizability across diverse human populations and clinical settings. I regularly encounter genomics studies where Manhattan plot associations identified in European populations fail to replicate in non-European ancestry groups, highlighting the importance of multi-ancestry approaches and population-specific analysis strategies for equitable genetic medicine development.
Effective Implementation in Genomics Research
Based on my extensive experience implementing Manhattan plots across diverse genomics research contexts, I have developed systematic approaches that maximize their analytical value and biological insight generation:
• Statistical Threshold Selection and Multiple Testing Control: Careful selection of genome-wide significance thresholds based on study design, population characteristics, and multiple testing burden proves critical for reliable genetic discovery while controlling false positive rates. I consistently recommend employing genome-wide significance thresholds appropriate for the specific study context, incorporating permutation-based significance assessment when standard Bonferroni correction may be overly conservative, and considering suggestive significance thresholds for hypothesis generation while maintaining appropriate statistical rigor for discovery claims.
• Genomic Annotation and Functional Context Integration: Sophisticated integration of genomic annotations, functional predictions, and biological pathway information transforms Manhattan plots from statistical summaries into comprehensive genetic analyses that connect association signals with functional consequences and therapeutic opportunities. In my functional genomics research, I routinely incorporate gene annotation tracks, regulatory element mapping, and pathway enrichment results that enable identification of biologically coherent association patterns while facilitating mechanistic hypothesis generation and experimental validation planning.
• Population Structure Control and Ancestry Analysis: Systematic approaches to population structure correction, ancestry inference, and demographic modeling prove essential for generating reliable genetic associations that can be appropriately interpreted across diverse human populations and clinical contexts. I frequently employ principal component analysis, admixture modeling, and kinship matrix corrections that account for population history effects while preserving genuine genetic association signals and enabling ancestry-specific effect size estimation.
• Multi-Phenotype Integration and Pleiotropy Assessment: Complex genetic research often requires Manhattan plot strategies that accommodate multiple related phenotypes, longitudinal measurements, or multi-trait analyses that reveal shared genetic architecture and pleiotropy patterns across related diseases or quantitative traits. In my experience with multi-phenotype GWAS studies, I recommend approaches that employ multivariate statistical methods, cross-trait genetic correlation analysis, and phenome-wide association studies that maximize discovery power while controlling for multiple phenotype testing burdens.
Real Examples from Leading Genomics Research
The following examples from our curated collection demonstrate how leading genetics researchers effectively implement Manhattan plots across diverse genomics contexts. Each plot represents peer-reviewed research from top-tier genetics journals, showcasing sophisticated association analysis approaches that advance genetic understanding.
Cardiovascular Disease Genetics
Genome-wide association meta-analysis revealing heart failure susceptibility loci - View full plot details
Cardiovascular genetics research demonstrates Manhattan plot excellence for complex disease association studies. The Nature Genetics publication investigating heart failure genetics through meta-analysis (DOI: 10.1038/s41588-024-02064-3) employs Manhattan plots to present genome-wide association results across multiple heart failure subtypes and population cohorts. The visualization effectively reveals novel susceptibility loci while demonstrating polygenic architecture patterns that inform therapeutic target identification and precision medicine approaches for cardiovascular disease prevention.
Regulatory Genomics and Protein QTL Analysis
Whole-genome sequencing identification of protein level regulatory variants - View full plot details
Functional genomics research showcases Manhattan plot applications for molecular phenotype mapping. The Nature Genetics publication investigating circulating protein QTLs (DOI: 10.1038/s41588-025-02095-4) uses Manhattan plots to present associations between rare genomic variants and protein abundance levels. The researchers effectively demonstrate how whole-genome sequencing enables identification of large-effect regulatory variants that influence protein concentrations, revealing functional variants missed by array-based approaches.
Population Genetics and Ancestry Studies
Comparative ancestry analysis of clonal hematopoiesis genetic associations - View full plot details
Population genetics research demonstrates sophisticated Manhattan plot implementation for ancestry-specific association studies. The Nature Genetics publication comparing Mexico City and UK Biobank populations (DOI: 10.1038/s41588-025-02085-6) employs Manhattan plots to present clonal hematopoiesis associations across different ancestry groups. The visualization reveals ancestry-specific genetic effects while identifying shared and population-specific susceptibility loci that inform equitable genetic medicine development.
Neuropsychiatric Genetics and Trans-Ancestry Studies
Trans-ancestry genome-wide depression study revealing 697 genetic associations - View full plot details
Neuropsychiatric genetics research provides examples of Manhattan plot excellence in trans-ancestry association studies. The Cell publication investigating depression genetics across multiple ancestry groups (DOI: 10.1016/j.cell.2024.12.002) uses Manhattan plots to present genome-wide association results across diverse populations. The researchers effectively demonstrate how trans-ancestry approaches enhance genetic discovery while revealing population-specific and shared genetic architecture patterns that inform therapeutic target identification.
Rare Disease Genetics and Family Studies
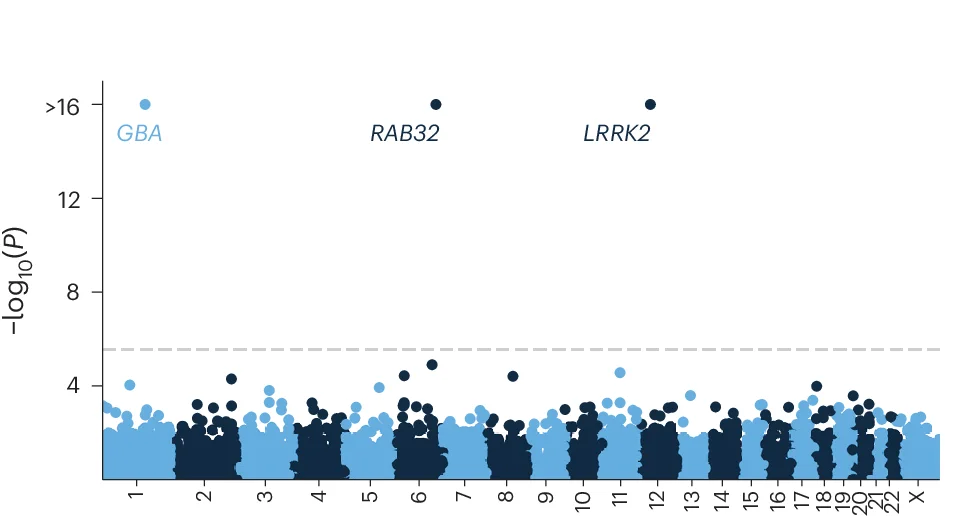
Systematic rare variant analysis identifying RAB32 in familial Parkinson disease - View full plot details
Rare disease genetics research showcases Manhattan plot applications for family-based association studies. The Nature Genetics publication investigating familial Parkinson's disease (DOI: 10.1038/s41588-024-01787-7) employs Manhattan plots to present rare variant association results across affected families. The visualization demonstrates how systematic rare variant analysis can identify novel susceptibility genes while accounting for family structure and inheritance patterns in Mendelian disease genetics.
Comparative Genomics and Evolutionary Studies
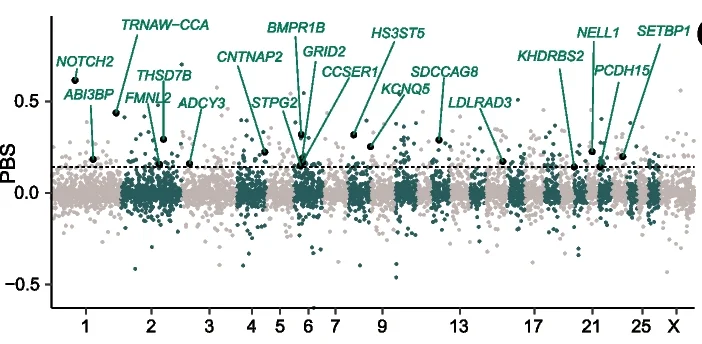
Structural variant landscape comparison revealing evolutionary signatures in sheep and goats - View full plot details
Evolutionary genetics research demonstrates advanced Manhattan plot implementation for comparative genomics studies. The Genome Biology publication investigating structural variant evolution (DOI: 10.1186/s13059-024-03288-6) uses Manhattan plots to present structural variant association patterns across different species and populations. The researchers effectively reveal convergent evolutionary signatures while identifying genomic regions under selection pressure that drive species adaptation and domestication traits.
Maximizing Genomics Discovery Impact
Based on my extensive experience implementing Manhattan plots across diverse genomics research contexts, several key principles consistently distinguish exceptional genetic discoveries from merely adequate association analyses:
• Functional Integration and Mechanistic Insight: The most effective Manhattan plot implementations combine genetic association results with functional annotation, expression data, and pathway analysis that transform statistical peaks into actionable biological understanding and therapeutic opportunities. I consistently recommend integrating eQTL data, chromatin state information, and protein interaction networks that enable connection of association signals with functional consequences and disease mechanisms.
• Multi-Population Validation and Equity Considerations: Context-appropriate Manhattan plot implementation must incorporate multi-ancestry validation, population-specific effect size assessment, and equity considerations that ensure genetic discoveries benefit diverse human populations rather than perpetuating healthcare disparities. In my collaborative population genetics research, I emphasize validation strategies that include diverse ancestry groups while accounting for population-specific linkage disequilibrium patterns and demographic history effects.
• Clinical Translation and Precision Medicine Integration: Future-oriented Manhattan plot implementation will increasingly incorporate clinical databases, drug target information, and precision medicine frameworks that facilitate translation of genetic associations into clinical applications and therapeutic development opportunities. However, the fundamental principles of appropriate statistical analysis, functional validation, and clinical relevance assessment will continue to determine the difference between meaningful genetic insight and statistical artifact identification.
Advancing Your Genomics Analysis Skills
The Manhattan plot examples featured in our curated collection represent the highest standards of genetic association analysis, drawn from publications in Nature Genetics, Cell, and other leading genetics journals. Each example demonstrates effective integration of statistical rigor with biological insight while advancing our understanding of complex genetic architecture through sophisticated association analysis approaches.
My analysis of thousands of Manhattan plot implementations across diverse genomics research contexts has reinforced their critical importance for genetic discovery and disease mechanism elucidation that drives precision medicine development. When implemented thoughtfully with attention to statistical accuracy, population diversity, and functional validation, Manhattan plots transform genome-wide association data into actionable genetic insights that advance scientific knowledge and clinical applications.
I encourage genomics researchers to explore our complete curated collection of Manhattan plot examples, where you can discover additional high-quality association analyses from cutting-edge genetics research across multiple disease areas and populations. Each plot includes comprehensive methodological documentation and genetic interpretation guidance, enabling you to adapt proven association analysis approaches to your own genomics research challenges and discovery objectives.
Want to explore more examples of professional Manhattan plot implementation from top-tier genetics publications? Check out our curated collection at: Manhattan Plot - featuring dozens of publication-quality genome-wide association analyses from Nature Genetics, Cell, and other leading genetics journals, each with complete statistical methodology details and functional validation examples.
Related Articles

Principal Component Analysis Visualization: PCA Plots in Genomics and Systems Biology
Master PCA plot creation for genomics and multi-omics research through real examples from Nature, Cell, and leading journals. Learn dimensionality reduction, variance explanation, and population structure analysis.

Differential Expression Analysis: Volcano Plot Mastery in Biological Research
Master volcano plot creation for genomics and proteomics research through real examples from Cell, Nature, and top biological journals. Learn fold-change analysis, significance thresholds, and interpretation.

Gene Expression Heatmaps: Pattern Recognition in Biological Research Publications
Master heatmap visualization for genomics and systems biology through real examples from Cell, Nature, and leading research journals. Learn clustering, color schemes, and pattern analysis.