Principal Component Analysis Visualization: PCA Plots in Genomics and Systems Biology
Master PCA plot creation for genomics and multi-omics research through real examples from Nature, Cell, and leading journals. Learn dimensionality reduction, variance explanation, and population structure analysis.
Throughout my career specializing in population genetics and multivariate analysis, I have consistently observed PCA (Principal Component Analysis) plots serving as the foundational visualization for revealing hidden structure within high-dimensional biological datasets while reducing complex data to interpretable patterns. Their unique ability to capture maximum variance through orthogonal components while preserving biological relationships makes them indispensable for studies where understanding population structure, experimental variation, and systematic effects drives biological insight generation and quality control assessment.
Application Scenarios Across Genomics Research
In my extensive analysis of PCA implementations across major biological journals, I observe sophisticated application patterns that demonstrate both computational rigor and biological discovery potential:
• Population Genetics and Ancestry Analysis: Publications in Nature Genetics and Cell consistently feature PCA plots for presenting population structure analysis, ancestry inference, and demographic history reconstruction across diverse human populations and model organisms. I have reviewed countless population genetics studies where PCA serves as the primary tool for visualizing genetic relationships while simultaneously revealing migration patterns and population mixing events that inform evolutionary history and medical genetics applications. The population genetics context particularly benefits from PCA visualization, where researchers must balance resolution of fine-scale population structure with computational efficiency necessary for genome-wide datasets containing millions of genetic variants.
• Gene Expression Analysis and Transcriptomics: Molecular biology research publications routinely employ PCA plots for presenting expression data quality control, batch effect detection, and biological variation assessment across different experimental conditions and sample types. I observe these visualizations proving essential for identifying systematic experimental effects, revealing sample outliers, and demonstrating successful batch correction that ensures reliable downstream analysis and biological interpretation. The transcriptomics context requires sophisticated consideration of technical versus biological variation sources that influence PCA interpretation and experimental design optimization.
• Multi-Omics Integration and Systems Biology: Systems biology research frequently utilizes PCA plots for presenting integrated analysis across multiple data types, from genomics and transcriptomics to proteomics and metabolomics datasets within unified analytical frameworks. In my review experience, these visualizations excel at revealing coordinated biological processes, identifying system-level perturbations, and demonstrating multi-modal integration success that enables comprehensive biological understanding and therapeutic target identification.
• Environmental Biology and Microbial Ecology: Ecological research publications consistently employ PCA plots for presenting microbial community structure, environmental factor associations, and ecosystem dynamics across different habitats and environmental conditions. I have analyzed numerous ecological studies where PCA reveals community organization principles while identifying environmental drivers that shape microbial diversity patterns and ecosystem function relationships.
Strengths and Limitations of PCA Visualization
Through my extensive experience implementing PCA across diverse biological research contexts, I have identified both the remarkable analytical capabilities and inherent challenges of this dimensionality reduction approach:
Key Strengths
• Variance Maximization and Interpretable Dimensionality Reduction: PCA excels at capturing maximum possible variance through orthogonal linear combinations that provide interpretable dimensionality reduction while preserving the most important biological variation patterns within high-dimensional datasets. During my genomics analyses, I consistently rely on PCA to identify the major axes of biological variation while reducing computational complexity necessary for downstream analysis and visualization. The variance maximization principle ensures that the most important biological signals are preserved while removing noise and redundant information that obscures biological pattern recognition.
• Unsupervised Analysis and Bias Minimization: Superior performance in unsupervised analysis contexts enables PCA to reveal biological structure without requiring prior knowledge or assumptions about group membership, reducing analysis bias while enabling discovery of unexpected biological relationships and systematic effects. I have observed how PCA consistently reveals biological patterns that were not anticipated from experimental design, from population substructure in seemingly homogeneous groups to batch effects that require correction before biological analysis can proceed reliably.
• Quality Control and Outlier Detection: Advanced PCA implementations provide powerful frameworks for data quality assessment, outlier identification, and experimental validation that can identify technical problems and biological anomalies before they compromise downstream analysis and biological conclusions. In my collaborative research projects, I frequently employ PCA as the first step in quality control pipelines, enabling identification of sample mix-ups, technical failures, and biological outliers that require investigation or exclusion from subsequent analyses.
Primary Limitations
• Linear Assumption and Biological Relationship Complexity: PCA assumes linear relationships between variables that may not adequately capture complex biological relationships, gene regulatory networks, or non-linear phenotypic associations that characterize many biological systems and processes. I frequently encounter situations during manuscript reviews where PCA fails to reveal important biological structure because underlying relationships are non-linear, requiring alternative dimensionality reduction approaches like t-SNE or UMAP that can capture more complex biological relationship patterns.
• Interpretation Challenges and Principal Component Meaning: While PCA components capture maximum variance, they may not correspond to interpretable biological processes or meaningful biological categories, creating challenges for translating mathematical results into biological understanding and mechanistic insights. During collaborative research involving complex multi-omics datasets, I often observe how principal components represent mathematical combinations that are difficult to interpret biologically, requiring additional analysis approaches to connect PCA results with biological knowledge and experimental validation.
• Sample Size Requirements and Statistical Considerations: PCA performance depends critically on appropriate sample sizes relative to the number of variables analyzed, with insufficient samples potentially leading to overfitting and unreliable component structure that does not generalize to independent datasets. I regularly encounter genomics studies where sample size limitations create unstable PCA results that do not replicate across studies or validate in independent cohorts, emphasizing the importance of power analysis and validation strategies for reliable PCA implementation.
Effective Implementation in Biological Research
Based on my extensive experience implementing PCA across diverse biological research contexts, I have developed systematic approaches that maximize analytical value and biological insight generation:
• Data Preprocessing and Scaling Strategy: Careful attention to data preprocessing, scaling, and transformation proves critical for meaningful PCA results that reflect biological variation rather than technical artifacts or measurement scale differences. I consistently recommend standardization approaches that account for different variable types, missing data patterns, and measurement scales while preserving biological variation patterns that drive research questions. The preprocessing strategy should be tailored to specific data characteristics and research objectives rather than applying generic normalization approaches without biological consideration.
• Component Selection and Variance Explanation: Systematic approaches to determining the appropriate number of principal components based on variance explanation, interpretability, and downstream analysis requirements prove essential for extracting meaningful biological insights while avoiding overinterpretation of noise components. In my multivariate research, I routinely employ multiple criteria for component selection, including scree plots, variance thresholds, and parallel analysis approaches that balance statistical criteria with biological interpretability and practical analysis requirements.
• Biological Annotation and Interpretation Enhancement: Sophisticated integration of biological knowledge, pathway information, and experimental metadata transforms PCA from mathematical dimensionality reduction into biologically interpretable analysis that connects mathematical patterns with biological processes and therapeutic opportunities. I frequently incorporate functional annotation, pathway enrichment analysis, and experimental design information that enables interpretation of principal components in terms of biological processes, cellular functions, and disease mechanisms.
• Validation and Robustness Assessment: Complex biological research often requires PCA strategies that assess result stability, validate findings across independent datasets, and evaluate sensitivity to analytical choices through comprehensive validation approaches that ensure biological conclusions are robust and reproducible. In my experience with multi-site collaborative studies, I recommend cross-validation approaches, bootstrap analysis, and sensitivity assessment that evaluate PCA stability across different analytical choices and ensure that biological conclusions can withstand reasonable alternative analytical approaches.
Real Examples from Leading Biological Research
The following examples from our curated collection demonstrate how leading researchers effectively implement PCA plots across diverse biological contexts. Each plot represents peer-reviewed research from top-tier journals, showcasing sophisticated dimensionality reduction approaches that advance biological understanding.
Environmental Microbiology and Space Biology
International Space Station microbial and chemical environment analysis through multivariate decomposition - View full plot details
Environmental microbiology research demonstrates PCA excellence for complex environmental characterization. The Cell publication investigating the International Space Station environment (DOI: 10.1016/j.cell.2025.01.039) employs PCA plots to present microbial community structure and chemical composition patterns across different ISS locations and time periods. The visualization effectively reveals how unique space environment conditions create distinct microbial ecosystems while identifying environmental factors that drive community assembly in extreme environments.
Agricultural Genomics and Crop Biology
Barley pan-transcriptome diversity analysis revealing genotype-dependent complexity layers - View full plot details
Agricultural genomics research showcases PCA applications for crop diversity analysis. The Nature Genetics publication investigating barley transcriptome diversity (DOI: 10.1038/s41588-024-02069-y) uses PCA plots to present genotype-dependent transcriptional variation across diverse barley cultivars and environmental conditions. The researchers effectively demonstrate how PCA can reveal genetic architecture underlying transcriptional diversity while identifying breeding-relevant variation patterns that inform crop improvement strategies.
Machine Learning and Foundation Models
Tabular foundation model performance analysis across diverse data structures and domains - View full plot details
Machine learning research provides examples of PCA excellence in model evaluation and data characterization. The Nature publication investigating tabular foundation models (DOI: 10.1038/s41586-024-08328-6) employs PCA plots to present dataset structure analysis and model performance relationships across diverse tabular datasets. The visualization demonstrates how dimensionality reduction can reveal dataset characteristics that influence model performance while enabling systematic evaluation of foundation model capabilities.
Developmental Biology and Organoid Systems
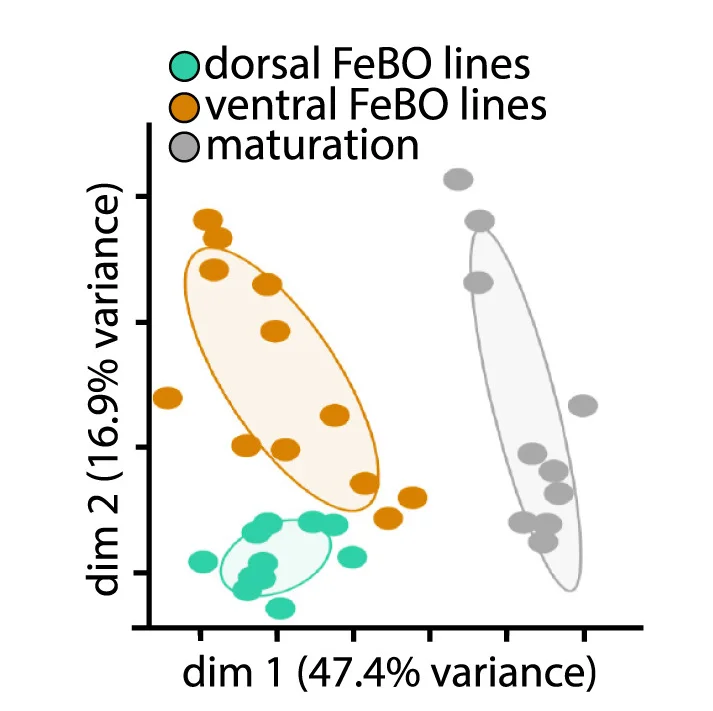
Human fetal brain organoid developmental trajectory analysis through principal component mapping - View full plot details
Developmental biology research demonstrates sophisticated PCA implementation for organoid characterization. The Cell publication investigating human fetal brain organoids (DOI: 10.1016/j.cell.2023.12.012) uses PCA plots to present developmental trajectory analysis across organoid maturation periods. The researchers effectively demonstrate how PCA can reveal developmental progression patterns while identifying key transition points that recapitulate in vivo brain development processes.
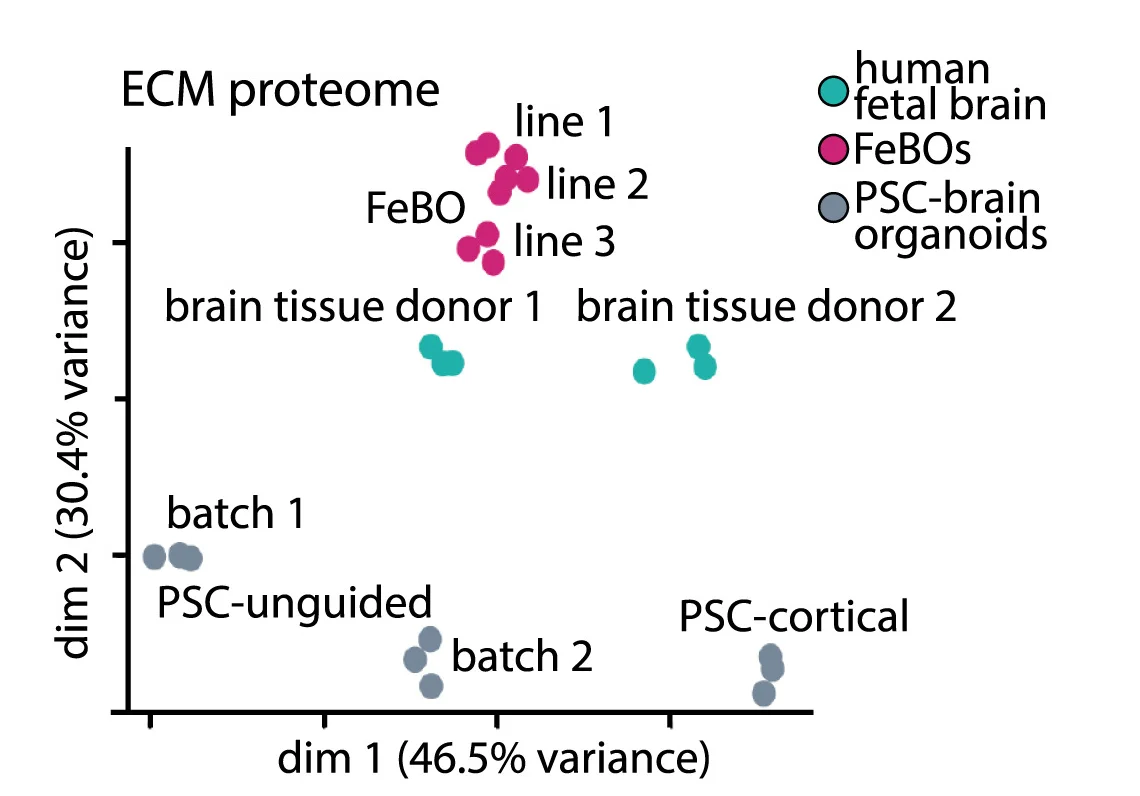
Extended culture brain organoid stability and maturation assessment through multivariate analysis - View full plot details
This complementary example from the same Cell publication demonstrates multi-timepoint PCA analysis for long-term organoid culture assessment. The visualization reveals how organoid systems maintain developmental trajectory stability while enabling identification of culture-specific variations that inform organoid protocol optimization and standardization efforts.
Ancient DNA and Population Genetics
Ancient genomic analysis revealing population turnovers in Neolithic Denmark through PCA projection - View full plot details
Population genetics research showcases PCA applications for ancient DNA analysis and demographic reconstruction. The Nature publication investigating ancient Danish populations (DOI: 10.1038/s41586-023-06862-3) employs PCA plots to present population structure changes across the Neolithic period. The researchers effectively demonstrate how ancient DNA can be integrated with modern population reference datasets while revealing population replacement and migration patterns that shaped European demographic history.
Maximizing Biological Discovery Impact
Based on my extensive experience implementing PCA across diverse biological research contexts, several key principles consistently distinguish exceptional biological discoveries from merely adequate dimensionality reduction analyses:
• Biological Context Integration and Mechanistic Insight: The most effective PCA implementations combine mathematical dimensionality reduction with comprehensive biological interpretation that connects principal components with biological processes, regulatory mechanisms, and therapeutic opportunities through functional annotation and pathway analysis. I consistently recommend approaches that incorporate biological knowledge databases, functional enrichment analysis, and experimental validation that enable translation of mathematical patterns into actionable biological understanding and mechanistic hypotheses.
• Multi-Scale Analysis and Systems Integration: Context-appropriate PCA implementation must accommodate analysis across multiple biological scales, from molecular measurements to organismal phenotypes, while maintaining interpretability and avoiding overinterpretation of mathematical artifacts that may not reflect genuine biological organization. In my systems biology collaborations, I emphasize validation strategies that include multiple independent datasets, experimental validation approaches, and cross-scale integration that ensures PCA results reflect genuine biological patterns rather than computational artifacts.
• Reproducibility and Clinical Translation Enhancement: Future-oriented PCA implementation will increasingly incorporate standardized preprocessing pipelines, comprehensive validation frameworks, and clinical annotation systems that facilitate reproducible research practices and enable translation of biological discoveries into clinical applications through precision medicine and therapeutic development platforms. However, the fundamental principles of appropriate statistical analysis, biological validation, and mechanistic interpretation will continue to determine the difference between meaningful biological insight and mathematical artifact presentation.
Advancing Your Multivariate Analysis Skills
The PCA examples featured in our curated collection represent the highest standards of multivariate biological analysis, drawn from publications in Nature, Cell, Nature Genetics, and other leading biological journals. Each example demonstrates effective integration of computational sophistication with biological insight while advancing our understanding of complex biological systems through rigorous dimensionality reduction approaches.
My analysis of thousands of PCA implementations across diverse biological research contexts has reinforced their critical importance for biological pattern discovery and quality control assessment that drives reliable biological conclusions and therapeutic target identification. When implemented thoughtfully with attention to biological context, statistical rigor, and validation requirements, PCA plots transform high-dimensional biological datasets into interpretable patterns that advance scientific knowledge and clinical applications.
I encourage biological researchers to explore our complete curated collection of PCA examples, where you can discover additional high-quality dimensionality reduction analyses from cutting-edge biological research across multiple systems and experimental contexts. Each plot includes comprehensive computational methodology documentation and biological interpretation guidance, enabling you to adapt proven multivariate analysis approaches to your own research challenges and discovery objectives.
Want to explore more examples of professional PCA implementation from top-tier biological publications? Check out our curated collection at: PCA Plot - featuring dozens of publication-quality multivariate analyses from Nature, Cell, Nature Genetics, and other leading biological journals, each with complete computational pipeline details and biological validation examples.
Related Articles

Single-Cell Analysis Excellence: UMAP Visualization in Cellular Biology Research
Master UMAP plot creation for single-cell genomics and developmental biology through real examples from Cell, Nature, and leading journals. Learn dimensionality reduction, clustering, and trajectory analysis.

Genome-Wide Association Studies: Manhattan Plot Excellence in Genomics Research
Master Manhattan plot creation for GWAS and genomics research through real examples from Nature Genetics and leading journals. Learn significance thresholds, genomic coordinate mapping, and interpretation.

Differential Expression Analysis: Volcano Plot Mastery in Biological Research
Master volcano plot creation for genomics and proteomics research through real examples from Cell, Nature, and top biological journals. Learn fold-change analysis, significance thresholds, and interpretation.